Imaging Biology Laboratory in Kumamoto University
We opened the Imaging Biology Laboratory (Higaki Laboratory) at Kumamoto University in the summer of 2017. Our laboratory members are interested in cytoskeleton dynamics in plant cell morphogenesis. We also develop image analysis methods from the perspective of experimental cell biologists.
Research history
Takumi Higaki, the PI of this laboratory, was a member of Hesezawa Laboratory, the University of Tokyo from 2004 to 2017, and has been conducting the following research. We accomplished time-sequential observations of plant actin filaments throughout cell cycle with GFP-tagged actin binding domain 2 of fimbrin that is an actin side-binding protein (Sano and Higaki et al. 2005 Plant J, Higaki et al. 2006 Plant Cell Physiol, Higaki et al. 2007 Plant Cell Physiol, Higaki et al. 2007 Curr Opin Plant Biol, Higaki et al. 2008 BMC Plant Biol). We also developed microscopic image analysis frameworks to quantitatively evaluate bundling, orientation, and density of actin filaments. Live cell imaging and the image processing techniques revealed that actin filaments were transiently bundled prior to stomatal opening, suggesting crucial roles of actin bundling in diurnal cycle-dependent stomatal opening (Higaki et al. 2010 Plant J). We also developed CARTA (Clustering-Aided Rapid Training Agent), which is computer software for rapid and accurate classification of the biomedical images based on active learning with iterative clustering (Kutsuna and Higaki et al. 2012 Nature Commun). Based on the CARTA methods, we also developed the computer-assisted system to detect the structures of interest, such as organelles (Higaki et al. 2015 Sci Rep). Furthermore, we tried to understand roles of microtubules in plant cell morphogenesis by an approach combining quantitative image processing and mathematical modeling. We proposed mathematical models to explain cell morphogenesis with microtubule distributions. The models successfully reproduce pattern formation observed in planta (Higaki et al. 2016 PLOS Comput Biol, Higaki et al. 2017 Plant Cell Physiol). In addition, we performed various collaborative works based on image processing techniques. In particular, we revealed intracellular dynamics in stomatal movements (Higaki et al. 2012 Sci Rep, Higaki et al. 2013 BMC Plant Biol, Hashimoto-Sugimoto and Higaki et al. 2013 Nature Commun, Higaki et al. 2014 Plant Cell Physiol, Higaki 2016 J Vis Exp), auxin response (Takahashi et al. 2016 Plant J), asymmetric cell division of zygotes (Kimata and Higaki et al. 2016 PNAS), plant immunity (Shimono and Higaki et al. 2016 PLOS One, Inada and Higaki et al. 2016 Plant Physiol, Inada and Higaki et al. 2016 J Plant Res), determination of cell division plane (Kojo and Higaki et al. 2013 Plant Cell Physiol), cell wall regeneration during protoplast culture (Yoneda et al. 2010 Plant J) and cytoplasmic streaming (Era et al. 2013 J Plant Res). Some of these works appeared on the cover of international journals as shown below.




Ongoing researches in this laboratory
In this laboratory, we are conducting various researches using plant cytoskeleton and image analysis techniques as keywords. For details, please refer to publication list. The following is a general outline of our research, but these are only examples and do not limit our future research.
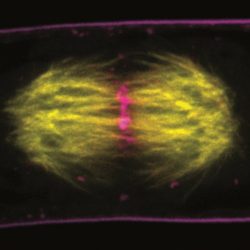
1. Cytoskeleton and cell morphogenesis in plants
Cytoskeleton is deeply involved in cell morphological changes through formation of higher-order structures such as networks and bundles. Using live cell imaging and microscopic image analysis techniques, we aim for understanding of organizations and roles of plant cytoskeleton roles in cell division, cell expansion, and cell death, in which plant cell undergoes dynamic morphological changes (PLOS Comput Biol 2016, Plant Physiol 2016, 2018ab, 2019, Plant J 2017, Plant Cell Physiol 2017, 2020).
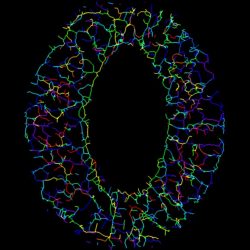
2. Stomatal development and movement
The stomata on the plant leaf and stem surfaces is essential for plant survival because they are responsible for gaseous exchange and transpiration. The stomatal density and aperture is appropriately regulated in response to environmental cues. We aim for understanding of the multi-scale spatiotemporal control mechanism of plant stomata (Plant J 2010, Sci Rep 2012, Nature Commun 2013, Plant Cell Physiol 2014, PLOS One 2013, 2016, Gene Cells 2020).
3. Image analysis tool development
We are developing technologies that are truly useful. We have developed the method for versatile biomedical image classification (Nature Commun 2012), detection of regions of interest in wide-area images (Sci Rep 2015), quantification of cytoskeleton bundling (Sci Rep 2020), and 3D reconstruction of plant architectures (Plant Cell Physiol 2021). We also support bioimage analysis technologies as collaborative research (PNAS 2016, 2019, Nature Plants 2018, Nature Commun 2020).

Contact
Takumi Higaki, Ph. D.
Kumamoto University, Kurokami South C7, Rm510
2-39-1, Kurokami, Chuou-ku, Kumamoto 860-8555, Japan
Tel:+81-96-342-3975
E-mail: thigaki{at}kumamoto-u.ac.jp


